Note
Go to the end to download the full example code.
Lowest Common Ancestors#
Compute and visualize LCA for node pairs
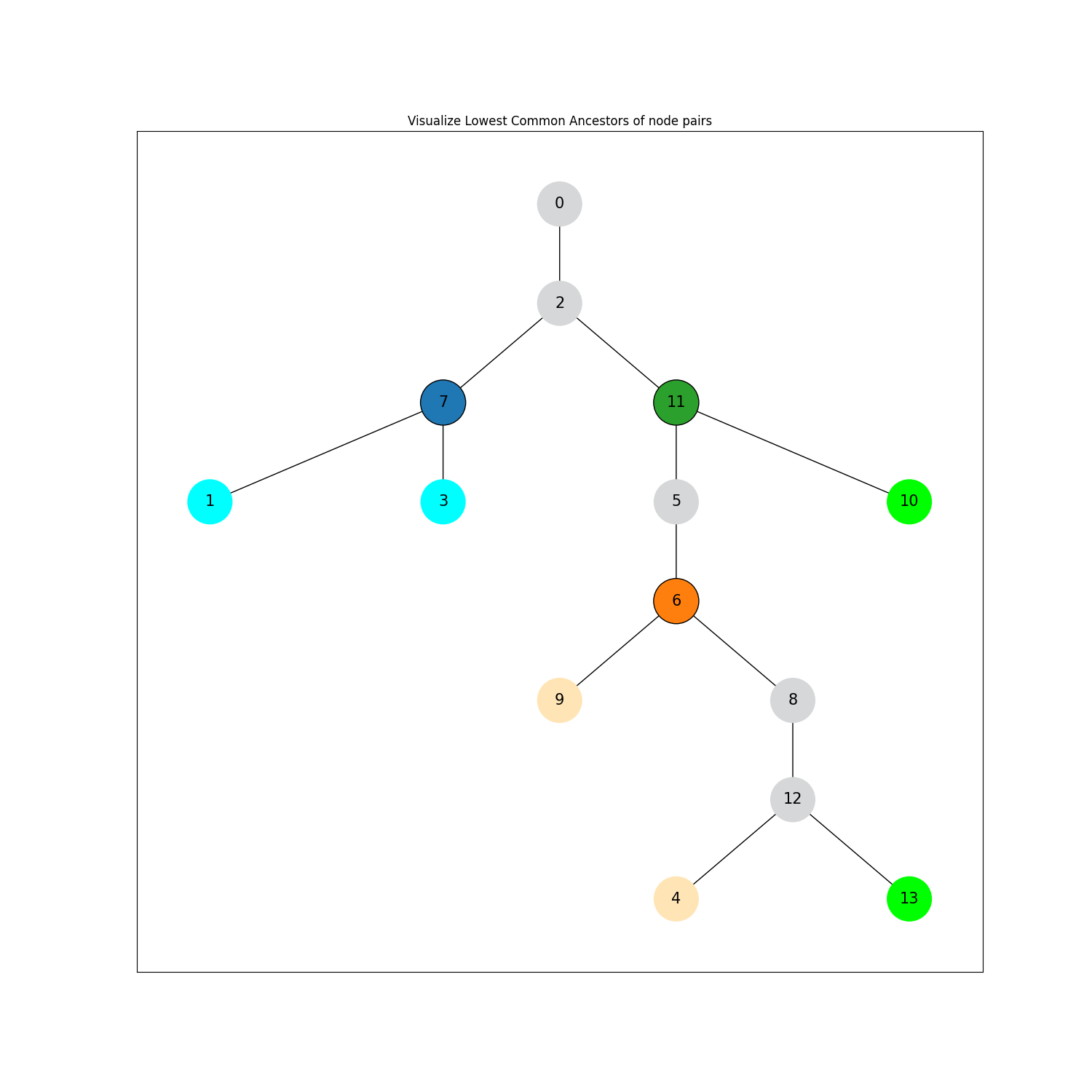
In a randomly generated directed tree, the lowest common ancestors are computed for certain node pairs. These node pairs and their LCA are then visualized with a chosen color scheme.
See also
- Lowest Common Ancestor
A more in-depth guide on lowest common ancestor algorithms in NetworkX.
import networkx as nx
import matplotlib.pyplot as plt
G = nx.DiGraph(
[
(0, 2),
(2, 7),
(2, 11),
(5, 6),
(6, 9),
(6, 8),
(7, 1),
(7, 3),
(8, 12),
(11, 10),
(11, 5),
(12, 4),
(12, 13),
]
)
pos = nx.nx_agraph.graphviz_layout(G, prog="dot")
# Compute lowest-common ancestors for certain node pairs
ancestors = list(nx.all_pairs_lowest_common_ancestor(G, ((1, 3), (4, 9), (13, 10))))
# Create node color and edge color lists
node_colors = ["#D5D7D8" for _ in G]
node_edge_colors = ["None" for _ in G]
node_seq = list(G.nodes)
clr_pairs = (("cyan", "tab:blue"), ("moccasin", "tab:orange"), ("lime", "tab:green"))
for (children, ancestor), (child_clr, anc_clr) in zip(ancestors, clr_pairs):
for c in children:
node_colors[node_seq.index(c)] = child_clr
node_colors[node_seq.index(ancestor)] = anc_clr
node_edge_colors[node_seq.index(ancestor)] = "black"
# Plot tree
plt.figure(figsize=(15, 15))
plt.title("Visualize Lowest Common Ancestors of node pairs")
nx.draw_networkx_nodes(
G, pos, node_color=node_colors, node_size=2000, edgecolors=node_edge_colors
)
nx.draw_networkx_edges(G, pos)
nx.draw_networkx_labels(G, pos, font_size=15)
plt.show()
Total running time of the script: (0 minutes 0.164 seconds)