Warning
This documents an unmaintained version of NetworkX. Please upgrade to a maintained version and see the current NetworkX documentation.
from_scipy_sparse_matrix¶
-
from_scipy_sparse_matrix(A, parallel_edges=False, create_using=None, edge_attribute='weight')[source]¶ Creates a new graph from an adjacency matrix given as a SciPy sparse matrix.
Parameters: - A (scipy sparse matrix) – An adjacency matrix representation of a graph
- parallel_edges (Boolean) – If this is
True,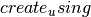 is a multigraph, and
is a multigraph, and 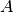 is an
integer matrix, then entry (i, j) in the matrix is interpreted as the
number of parallel edges joining vertices i and j in the graph. If it
is
is an
integer matrix, then entry (i, j) in the matrix is interpreted as the
number of parallel edges joining vertices i and j in the graph. If it
is False, then the entries in the adjacency matrix are interpreted as the weight of a single edge joining the vertices. - create_using (NetworkX graph) – Use specified graph for result. The default is Graph()
- edge_attribute (string) – Name of edge attribute to store matrix numeric value. The data will have the same type as the matrix entry (int, float, (real,imag)).
Notes
If
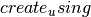 is an instance of
is an instance of networkx.MultiGraphornetworkx.MultiDiGraph,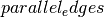 is
is True, and the entries of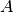 are of type
are of type int, then this function returns a multigraph (of the same type as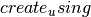 ) with parallel edges. In this case,
) with parallel edges. In this case,
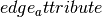 will be ignored.
will be ignored.If
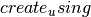 is an undirected multigraph, then only the edges
indicated by the upper triangle of the matrix
is an undirected multigraph, then only the edges
indicated by the upper triangle of the matrix 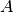 will be added to the
graph.
will be added to the
graph.Examples
>>> import scipy.sparse >>> A = scipy.sparse.eye(2,2,1) >>> G = nx.from_scipy_sparse_matrix(A)
If
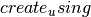 is a multigraph and the matrix has only integer entries,
the entries will be interpreted as weighted edges joining the vertices
(without creating parallel edges):
is a multigraph and the matrix has only integer entries,
the entries will be interpreted as weighted edges joining the vertices
(without creating parallel edges):>>> import scipy >>> A = scipy.sparse.csr_matrix([[1, 1], [1, 2]]) >>> G = nx.from_scipy_sparse_matrix(A, create_using=nx.MultiGraph()) >>> G[1][1] {0: {'weight': 2}}
If
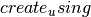 is a multigraph and the matrix has only integer entries
but
is a multigraph and the matrix has only integer entries
but 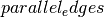 is
is True, then the entries will be interpreted as the number of parallel edges joining those two vertices:>>> import scipy >>> A = scipy.sparse.csr_matrix([[1, 1], [1, 2]]) >>> G = nx.from_scipy_sparse_matrix(A, parallel_edges=True, ... create_using=nx.MultiGraph()) >>> G[1][1] {0: {'weight': 1}, 1: {'weight': 1}}