networkx.convert.to_scipy_sparse_matrix¶
- networkx.convert.to_scipy_sparse_matrix(G, nodelist=None, dtype=None)¶
Return the graph adjacency matrix as a SciPy sparse matrix.
Parameters : G : graph
The NetworkX graph used to construct the NumPy matrix.
nodelist : list, optional
The rows and columns are ordered according to the nodes in
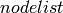 .
If
.
If 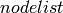 is None, then the ordering is produced by G.nodes().
is None, then the ordering is produced by G.nodes().dtype : NumPy data-type, optional
A valid NumPy dtype used to initialize the array. If None, then the NumPy default is used.
Returns : M : SciPy sparse matrix
Graph adjacency matrix.
Notes
The matrix entries are populated using the ‘weight’ edge attribute. When an edge does not have the ‘weight’ attribute, the value of the entry is 1.
For multiple edges the matrix values are the sums of the edge weights.
When
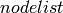 does not contain every node in
does not contain every node in 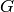 , the matrix is built
from the subgraph of
, the matrix is built
from the subgraph of 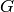 that is induced by the nodes in
that is induced by the nodes in 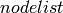 .
.Uses lil_matrix format. To convert to other formats see the documentation for scipy.sparse.
Examples
>>> G = nx.MultiDiGraph() >>> G.add_edge(0,1,weight=2) >>> G.add_edge(1,0) >>> G.add_edge(2,2,weight=3) >>> G.add_edge(2,2) >>> S = nx.to_scipy_sparse_matrix(G, nodelist=[0,1,2]) >>> S.todense() matrix([[ 0., 2., 0.], [ 1., 0., 0.], [ 0., 0., 4.]])