networkx.clustering¶
- clustering(G, nbunch=None, with_labels=False, weights=False)¶
Compute the clustering coefficient for nodes.
For each node find the fraction of possible triangles that exist,
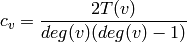
where
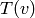 is the number of triangles through node
is the number of triangles through node  .
.Parameters: G : graph
A networkx graph
nbunch : container of nodes, optional
Limit to specified nodes. Default is entire graph.
with_labels: bool, optional :
If True return a dictionary keyed by node label.
weights : bool, optional
If True return fraction of connected triples as dictionary
Returns: out : float, list, dictionary or tuple of dictionaries
Clustering coefficient at specified nodes
Notes
The weights are the fraction of connected triples in the graph which include the keyed node. Ths is useful for computing transitivity.
Self loops are ignored.
Examples
>>> G=nx.complete_graph(5) >>> print nx.clustering(G,0) 1.0 >>> print nx.clustering(G,with_labels=True) {0: 1.0, 1: 1.0, 2: 1.0, 3: 1.0, 4: 1.0}